Background
The degree to which diet reproducibly alters the human and mouse gut microbiota remains unclear. Through a unbiased subject-level meta-analysis framework, we re-analyzed 27 studies including 1101 samples from rodents and humans. We demonstrate reproducible changes in gut microbial community structure; however, reduction in alpha diversity is not a consistent feature of HFD feeding. Finer taxonomic analysis revealed that Lactococcus spp. are the most reproducible signal of HFD, which we experimentally demonstrate are common dietary contaminants. We employed machine learning to define signatures capable of predicting the dietary intake of mice and demonstrate that phylogenetic and gene-centric transformations of this model are capable of translating to humans. Together, these results demonstrate the utility of microbiome meta-analyses in identifying robust bacterial signals for mechanistic studies and creates a framework for the routine meta-analysis of microbiome studies in preclinical models.
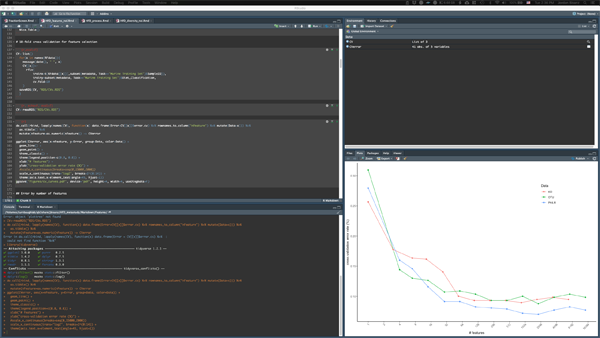
This page contains links to the R Markdown documents that contain the line by line commands that recapitulate all figure and analysis included in the manuscript as well as processed versions of data useful for reanalysis.